Microsoft and University of Washington (UW) researchers have demonstrated the ability to use synthetic DNA as a form of archival storage for data.
If the technology can be made robust enough for mainstream use, it would be possible to take a Walmart-sized supercenter filled with today's highest capacity data storage devices and shrink it to the size of a sugar cube, the researchers said.
"We think the time is ripe to consider DNA-based storage seriously and explore system designs and architectural implications," the researchers wrote in their paper.
The research team was able to successfully encode digital data from four image files into the nucleotide sequences of synthetic DNA snippets. More significantly, they were also able to reverse that process — retrieving the correct sequences from a larger pool of DNA and reconstructing the images without losing a single byte of information.

UW computer science and engineering research scientists mix DNA samples for storage. Each tube contains a digital file, which might be a picture of a cat or a Tchaikovsky symphony.
Another experiment demonstrated the ability to encode and retrieve data that authenticates archival video files from the UW's "Voices from the Rwanda Tribunal" project that contain 49 video interviews with judges, lawyers and other personnel from the Rwandan war crime tribunal.
"Life has produced this fantastic molecule called DNA that efficiently stores all kinds of information about your genes and how a living system works — it's very, very compact and very durable," Luis Ceze, UW associate professor of computer science and engineering and co-author of the research paper, said in a statement.
"We're essentially repurposing it to store digital data — pictures, videos, documents — in a manageable way for hundreds or thousands of years," he added.
Research into DNA data storage has progressed rapidly. In 1999, DNA-based storage involved encoding and recovering a 23-character message.
By 2013, scientists from U.K.-based EMBL-European Bioinformatics Institute claimed they'd encoded an .mp3 of Martin Luther King's "I Have a Dream" speech in DNA.
The encoding method makes it possible to store at least 100 million hours of high-definition video in about a cup of DNA, the researchers said in a paper published in the journal Nature.
According to the U.K. researchers, data stored in strands of DNA can last for tens of thousands of years.
Reading DNA is fairly straightforward, but writing it has been a major hurdle. There are two challenges: First, using current methods, it is only possible to manufacture DNA in short strings. Secondly, both writing and reading DNA are prone to errors, particularly when the same DNA letter is repeated.

Three image files were synthesized and sequenced for the DNA data storage experiments.
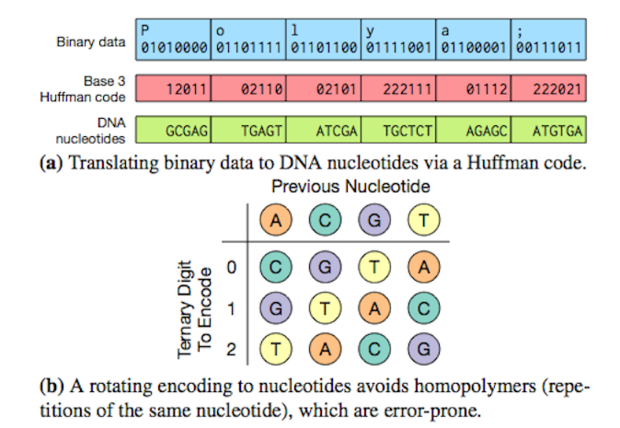
The Microsoft and UW researchers said they developed "a novel approach" to convert the long strings of ones and zeroes in digital data into the four basic building blocks of DNA sequences — adenine, guanine, cytosine and thymine - represented as As, Gs, Cs and Ts.
To access the stored data, the researchers encode the equivalent of zip codes and street addresses into the DNA sequences. Polymerase Chain Reaction (PCR) techniques — commonly used in molecular biology — help them more easily identify the zip codes they are looking for.
Using DNA sequencing techniques, the researchers can then "read" the data and convert it back to a video, image or document file by using the street addresses to reorder the data.
"How you go from ones and zeroes to As, Gs, Cs and Ts really matters because if you use a smart approach, you can make it very dense and you don't get a lot of errors," said co-author Georg Seelig, a UW associate professor of electrical engineering and of computer science and engineering.
The Microsoft and UW researchers announced their breakthrough at the ACM International Conference on Architectural Support for Programming Languages and Operating Systems.
"DNA is an attractive possibility," the researchers said, because it is extremely dense, with a theoretical limit that is eight orders of magnitude denser than tape. Magnetic tape technology can store as much as 185TB on a single cartridge that can fit in the palm of your hand.
The Microsoft and UW researchers also confirmed synthetic DNA's longevity, saying it has a half-life of more than 500 years in harsh environments. Tape cartridges have a lifespan of 10 to 30 years and hard disk drives are rated to last three to five years, the researchers noted.
The U.S. researchers emphasized the need for a more dense archival medium as all the data contained in our computers, historic archives, movies, photos and businesses systems and mobile devices worldwide is expected to hit 44 trillion gigabytes by 2020, according to The Digital Universe research paper from IDC and EMC.
"That's a 10-fold increase compared to 2013, and will represent enough data to fill more than six stacks of computer tablets stretching to the moon. While not all of that information needs to be saved, the world is producing data faster than the capacity to store it," the researchers said in their paper.
A DNA storage system still has problems that must overcome before its ready for commercial use. First, DNA synthesis and sequencing is far from perfect, with error rates on the order of 1% per nucleotide. A key aspect of DNA storage will be to devise appropriate encoding schemes that can tolerate errors by adding redundancy.
Additionally, randomly accessing data in DNA-based storage is problematic, resulting in overall read latency that is much longer than write latency. The current efforts have provided only large-block access; to read even a single byte from storage, the entire DNA pool must be sequenced and decoded.
The scientists have proposed ways to improve random access by using a polymerase chain reaction (PCR) to amplify only the desired data, biasing sequencing towards it. That both accelerates reads and ensures that an entire DNA pool need not be sequenced.
"This is an example where we're borrowing something from nature — DNA — to store information," Ceze stated. "But we're using something we know from computers — how to correct memory errors — and applying that back to nature."
This story, "Scientists could use DNA to shrink a data center into a sugar cube" was originally published by Computerworld.